Euler characteristic curves and profiles
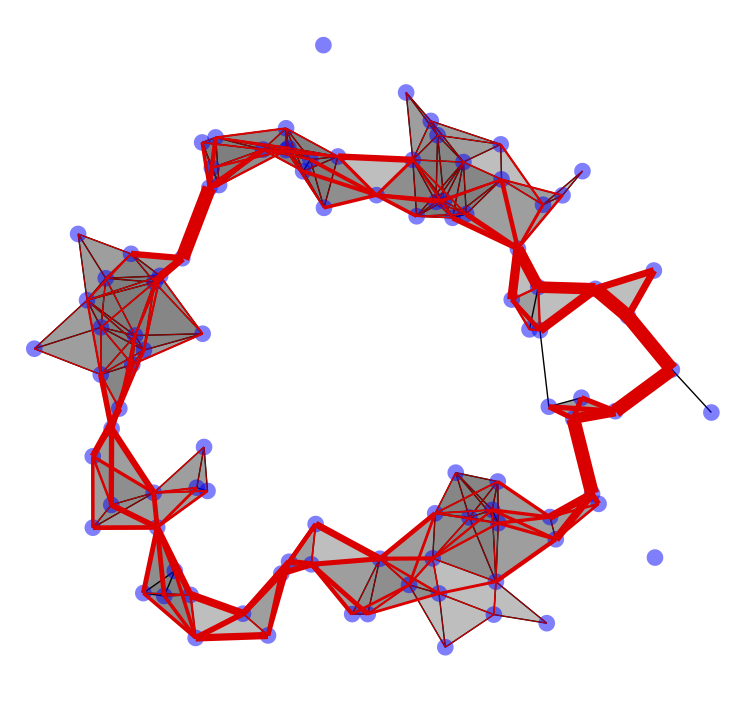
The Euler characteristic is probably one of the oldest topological invariants, in this work we analyze it in the context of filtered data. By building a filtered cell complex on top of it and tracking the evolution of its Euler characteristic at different scales we obtain an Euler characteristic curve, which can be used as a topological feature.
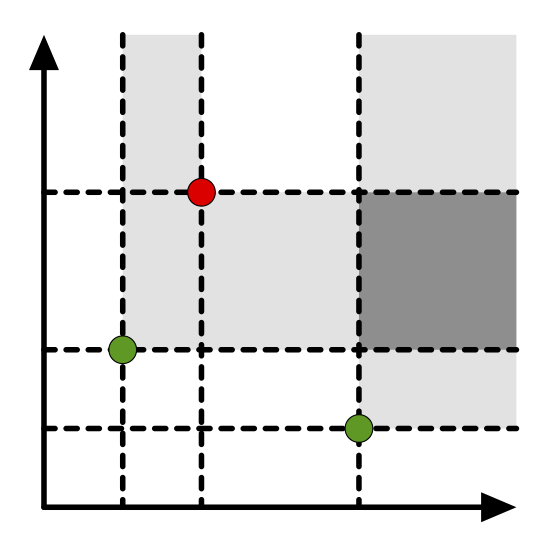
We prove a novel stability result, bounding the distance between two Euler characteristic curves obtained from two filtered complexes with the Wasserstein distance between their persistence diagrams, a widely studied distance in TDA for which many stability results are known. Moreover, we generalize the Euler characteristic curve to complexes whose filtrations take values in a general poset, thus lifting it in the realm of multiparameter persistence. We call this generalization Euler characteristic profile.
Our stability results justify the use of Euler characteristic curves and profiles as data descriptors. In order to do so, we provide distributed algorithms to compute them in an efficient way for different types of data, leveraging the combinatorial structure of different types of cell complexes, namely Vietoris-Rips simplicial complexes and cubical complexes. Our proposed algorithms are fully parallelizable and can be executed in a streaming fashion, without the need to load the entire input into memory all at once. This paves the way for TDA applications in big data scenarios.